pytdc1.1.15
Published
Therapeutics Commons
pip install pytdc
Package Downloads
Authors
Project URLs
Requires Python
Dependencies
- accelerate
==0.33.0 - dataclasses
<1.0,>=0.6 - datasets
<2.20.0 - evaluate
==0.4.2 - fuzzywuzzy
<1.0,>=0.18.0 - huggingface_hub
<1.0,>=0.20.3 - numpy
<2.0.0,>=1.26.4 - openpyxl
<4.0.0,>=3.0.10 - pandas
<3.0.0,>=2.1.4 - requests
<3.0.0,>=2.31.0 - scikit-learn
>=1.2.2 - seaborn
<1.0.0,>=0.12.2 - tqdm
<5.0.0,>=4.65.0 - transformers
<4.51.0,>=4.43.0 - cellxgene-census
==1.15.0 - gget
<1.0.0,>=0.28.4 - pydantic
<3.0.0,>=2.6.3 - rdkit
<2024.3.1,>=2023.9.5 - tiledbsoma
<2.0.0,>=1.7.2

Website | NeurIPS 2024 AIDrugX Paper | Nature Chemical Biology 2022 Paper | NeurIPS 2021 Paper | Long Paper | Slack | TDC Mailing List | TDC Documentation | Contribution Guidelines | [PyTDC Webpage]
(NEW) Introducing PyTDC
Existing biomedical benchmarks do not provide end-to-end infrastructure for training, evaluation, and inference of models that integrate multimodal biological data and a broad range of machine learning tasks in therapeutics. We present PyTDC, an open-source machine-learning platform providing streamlined training, evaluation, and inference software for multimodal biological AI models. PyTDC unifies distributed, heterogeneous, continuously updated data sources and model weights and standardizes benchmarking and inference endpoints.
The components of PyTDC include:
- A collection of multimodal, continually updated heterogeneous data sources is unified under the introduced "API-first-dataset" architecture. Inspired by API-first design, this microservice architecture uses the model-view-controller design pattern to enable multimodal data views.
- PyTDC presents open-source model retrieval and deployment software that streamlines AI inferencing and exposes state-of-the-art, research-ready models and training setups for biomedical representation learning models across modalities.
- We integrate single-cell analysis with multimodal machine learning in therapeutics via the introduction of contextualized tasks.
.png)
Built on the Therapeutic Data Commons (TDC)
Artificial intelligence is poised to reshape therapeutic science. Therapeutics Data Commons is a coordinated initiative to access and evaluate artificial intelligence capability across therapeutic modalities and stages of discovery. It supports the development of AI methods and aims to establish the foundation of which AI methods are most suitable for drug discovery applications and why.
Researchers across disciplines can use TDC for numerous applications. AI-solvable tasks, AI-ready datasets, and curated benchmarks in TDC serve as a meeting point between biochemical and AI scientists. TDC facilitates algorithmic and scientific advances and accelerates machine learning method development, validation, and transition into biomedical and clinical implementation.
TDC is an open-science initiative. We welcome contributions from the community.
Key (Py)TDC Presentations and Publications
[0] Western Bioinformatics Seminar Series: Alejandro Velez-Arce, "Signals in the Cells: Multimodal and Contextualized Machine Learning Foundations for Therapeutics." [Event] [Seminar] [Slides]
[1] Velez-Arce, Huang, Li, Lin, et al., Signals in the Cells: Multimodal and Contextualized Machine Learning Foundations for Therapeutics, NeurIPS AIDrugX, 2024 [Paper] [Slides] [Webpage]
[2] Huang, Fu, Gao, et al., Artificial Intelligence Foundation for Therapeutic Science, Nature Chemical Biology, 2022 [Paper]
[3] Huang, Fu, Gao, et al., Therapeutics Data Commons: Machine Learning Datasets and Tasks for Drug Discovery and Development, NeurIPS 2021 [Paper] [Poster]
[4] Huang et al., Benchmarking Molecular Machine Learning in Therapeutics Data Commons, ELLIS ML4Molecules 2021 [Paper] [Slides]
[5] Huang et al., Therapeutics Data Commons: Machine Learning Datasets and Tasks for Drug Discovery and Development, Baylearn 2021 [Slides] [Poster]
[6] Huang, Fu, Gao et al., Therapeutics Data Commons, NSF-Harvard Symposium on Drugs for Future Pandemics 2020 [#futuretx20] [Slides] [Video]
[7] TDC User Group Meetup, Jan 2022 [Agenda]
[8] Zitnik, Machine Learning to Translate the Cancer Genome and Epigenome Session, AACR Annual Meeting 2022, Apr 2022
[9] Zitnik, Few-Shot Learning for Network Biology, Keynote at KDD Workshop on Data Mining in Bioinformatics
[10] Zitnik, Actionable machine learning for drug discovery and development, Broad Institute, Models, Inference & Algorithms Seminar, 2021
[11] Zitnik, Graph Neural Networks for Biomedical Data, Machine Learning in Computational Biology, 2020
[12] Zitnik, Graph Neural Networks for Identifying COVID-19 Drug Repurposing Opportunities, MIT AI Cures, 2020
Unique Features of TDC
- Diverse areas of therapeutics development: TDC covers a wide range of learning tasks, including target discovery, activity screening, efficacy, safety, and manufacturing across biomedical products, including small molecules, antibodies, and vaccines.
- Ready-to-use datasets: TDC is minimally dependent on external packages. Any TDC dataset can be retrieved using only three lines of code.
- Data functions: TDC provides extensive data functions, including data evaluators, meaningful data splits, data processors, and molecule generation oracles.
- Leaderboards: TDC provides benchmarks for fair model comparison and systematic model development and evaluation.
- Open-source initiative: TDC is an open-source initiative. If you'd like to get involved, please don't hesitate to let us know.
See here for the latest updates in TDC!
Installation
Using pip
To install the core environment dependencies of TDC, use pip:
pip install PyTDC
Note: TDC is in the beta release. Please update your local copy regularly by
pip install PyTDC --upgrade
The core data loaders are lightweight with minimum dependency on external packages:
numpy, pandas, tqdm, scikit-learn, fuzzywuzzy, seaborn
Tutorials
We provide tutorials to get started with TDC:
| Name | Description |
|---|---|
| 101 | Introduce TDC Data Loaders |
| 102 | Introduce TDC Data Functions |
| 103.1 | Walk through TDC Small Molecule Datasets |
| 103.2 | Walk through TDC Biologics Datasets |
| 104 | Generate 21 ADME ML Predictors with 15 Lines of Code |
| 105 | Molecule Generation Oracles |
| 106 | Benchmark submission |
| DGL | Demo presented at DGL GNN User Group Meeting |
| U1.1 | Demo presented at first TDC User Group Meetup |
| U1.2 | Demo presented at first TDC User Group Meetup |
| 201 | TDC-2 Resource and Multi-modal Single-Cell API |
| 202 | TDC-2 Resource and PrimeKG |
| 203 | TDC-2 Resource and External APIs |
| 204 | TDC-2 Model Hub |
| 205 | TDC-2 Molecular Property Cliff Prediction Task |
Design of TDC
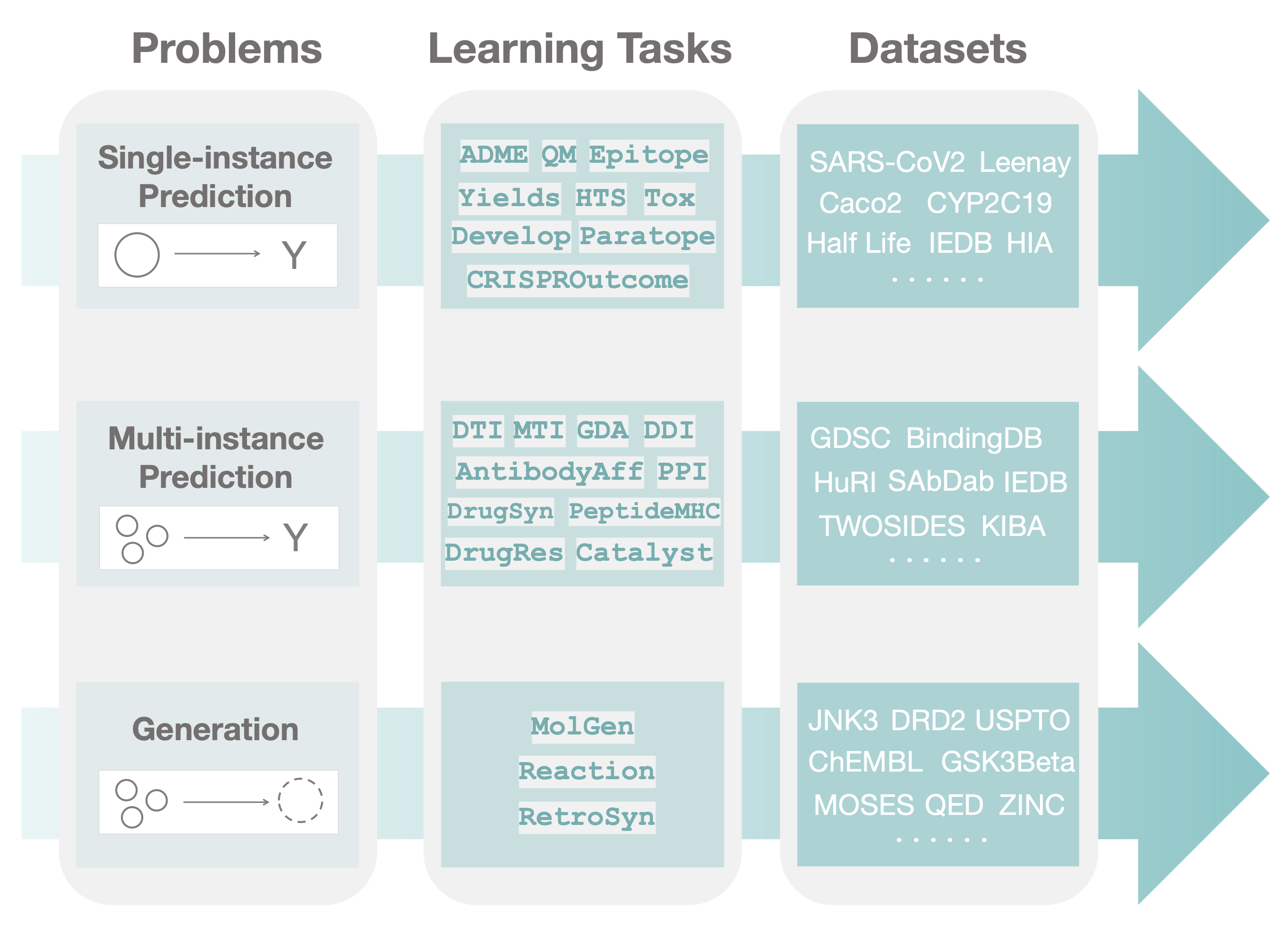
TDC has a unique three-tiered hierarchical structure, which to our knowledge, is the first attempt at systematically organizing machine learning for therapeutics. We organize TDC into three distinct problems. For each problem, we provide a collection of learning tasks. Finally, for each task, we provide a series of datasets.
In the first tier, after observing a large set of therapeutics tasks, we categorize and abstract out three major areas (i.e., problems) where machine learning can facilitate scientific advances, namely, single-instance prediction, multi-instance prediction, and generation:
- Single-instance prediction
single_pred: Prediction of property given individual biomedical entity. - Multi-instance prediction
multi_pred: Prediction of property given multiple biomedical entities. - Generation
generation: Generation of new desirable biomedical entities.

The second tier in the TDC structure is organized into learning tasks. Improvement in these tasks can result in numerous applications, including identifying personalized combinatorial therapies, designing novel classes of antibodies, improving disease diagnosis, and finding new cures for emerging diseases.
Finally, in the third tier of TDC, each task is instantiated via multiple datasets. For each dataset, we provide several splits into training, validation, and test sets to simulate the type of understanding and generalization (e.g., the model's ability to generalize to entirely unseen compounds or to granularly resolve patient response to a polytherapy) needed for transition into production and clinical implementation.
TDC Data Loaders
TDC provides a collection of workflows with intuitive, high-level APIs for both beginners and experts to create machine learning models in Python. Building off the modularized "Problem -- Learning Task -- Data Set" structure (see above) in TDC, we provide a three-layer API to access any learning task and dataset. This hierarchical API design allows us to easily incorporate new tasks and datasets.
For a concrete example, to obtain the HIA dataset from the ADME therapeutic learning task in the single-instance prediction problem:
from tdc.single_pred import ADME
data = ADME(name = 'HIA_Hou')
# split into train/val/test with scaffold split methods
split = data.get_split(method = 'scaffold')
# get the entire data in the various formats
data.get_data(format = 'df')
You can see all the datasets that belong to a task as follows:
from tdc.utils import retrieve_dataset_names
retrieve_dataset_names('ADME')
See all therapeutic tasks and datasets on the TDC website!
TDC Data Functions
Dataset Splits
To retrieve the training/validation/test dataset split, you could type
data = X(name = Y)
data.get_split(seed = 42)
# {'train': df_train, 'val': df_val, 'test': df_test}
You can specify the function's splitting method, random seed, and split fractions by, e.g., data.get_split(method = 'scaffold', seed = 1, frac = [0.7, 0.1, 0.2]). Check the data split page for details.
Strategies for Model Evaluation
We provide various evaluation metrics for the tasks in TDC, described in model evaluation page on the website. For example, to use metric ROC-AUC, you could type
from tdc import Evaluator
evaluator = Evaluator(name = 'ROC-AUC')
score = evaluator(y_true, y_pred)
Data Processing
TDC provides numerous data processing functions, including label transformation, data balancing, pairing data to PyG/DGL graphs, negative sampling, database querying, and so on. For function usage, see our data processing page on the TDC website.
Molecule Generation Oracles
For molecule generation tasks, we provide 10+ oracles for both goal-oriented and distribution learning. For detailed usage of each oracle, please have a look at the oracle page on the website. For example, we want to retrieve the GSK3Beta oracle:
from tdc import Oracle
oracle = Oracle(name = 'GSK3B')
oracle(['CC(C)(C)....'
'C[C@@H]1....',
'CCNC(=O)....',
'C[C@@H]1....'])
# [0.03, 0.02, 0.0, 0.1]
TDC Leaderboards
Every dataset in TDC is a benchmark, and we provide training/validation and test sets for it, together with data splits and performance evaluation metrics. To participate in the leaderboard for a specific benchmark, follow these steps:
-
Use the TDC benchmark data loader to retrieve the benchmark.
-
Use training and/or validation set to train your model.
-
Use the TDC model evaluator to calculate your model's performance on the test set.
-
Submit the test set performance to a TDC leaderboard.
As many datasets share a therapeutics theme, we organize benchmarks into meaningfully defined groups, which we refer to as benchmark groups. Datasets and tasks within a benchmark group are carefully curated and centered around a theme (for example, TDC contains a benchmark group to support ML predictions of the ADMET properties). While every benchmark group consists of multiple benchmarks, it is possible to separately submit results for each benchmark. Here is the code framework to access the benchmarks:
from tdc import BenchmarkGroup
group = BenchmarkGroup(name = 'ADMET_Group', path = 'data/')
predictions_list = []
for seed in [1, 2, 3, 4, 5]:
benchmark = group.get('Caco2_Wang')
# all benchmark names in a benchmark group are stored in group.dataset_names
predictions = {}
name = benchmark['name']
train_val, test = benchmark['train_val'], benchmark['test']
train, valid = group.get_train_valid_split(benchmark = name, split_type = 'default', seed = seed)
# --------------------------------------------- #
# Train your model using train, valid, test #
# Save test prediction in y_pred_test variable #
# --------------------------------------------- #
predictions[name] = y_pred_test
predictions_list.append(predictions)
results = group.evaluate_many(predictions_list)
# {'caco2_wang': [6.328, 0.101]}
For more information, visit here.
Cite Us
If you find Therapeutics Data Commons useful, cite our NeurIPS'24 AIDrugX paper, our NeurIPS paper, and Nature Chemical Biology paper :
@inproceedings{
velez-arce2024signals,
title={Signals in the Cells: Multimodal and Contextualized Machine Learning Foundations for Therapeutics},
author={Alejandro Velez-Arce and Xiang Lin and Kexin Huang and Michelle M Li and Wenhao Gao and Bradley Pentelute and Tianfan Fu and Manolis Kellis and Marinka Zitnik},
booktitle={NeurIPS 2024 Workshop on AI for New Drug Modalities},
year={2024},
url={https://openreview.net/forum?id=kL8dlYp6IM}
}
@article{Huang2021tdc,
title={Therapeutics Data Commons: Machine Learning Datasets and Tasks for Drug Discovery and Development},
author={Huang, Kexin and Fu, Tianfan and Gao, Wenhao and Zhao, Yue and Roohani, Yusuf and Leskovec, Jure and Coley,
Connor W and Xiao, Cao and Sun, Jimeng and Zitnik, Marinka},
journal={Proceedings of Neural Information Processing Systems, NeurIPS Datasets and Benchmarks},
year={2021}
}
@article{Huang2022artificial,
title={Artificial intelligence foundation for therapeutic science},
author={Huang, Kexin and Fu, Tianfan and Gao, Wenhao and Zhao, Yue and Roohani, Yusuf and Leskovec, Jure and Coley,
Connor W and Xiao, Cao and Sun, Jimeng and Zitnik, Marinka},
journal={Nature Chemical Biology},
year={2022}
}
TDC is built on top of other open-sourced projects. Additionally, please cite the original work if you used these datasets/functions in your research. You can find the original paper for the function/dataset on the website.
Contribute
TDC is a community-driven and open-science initiative. To get involved, join our Slack Workspace and check out the contribution guide!
Contact
Reach us at [email protected] or open a GitHub issue.
Data Server
Many TDC datasets are hosted on Harvard Dataverse with the following persistent identifier https://doi.org/10.7910/DVN/21LKWG. When Dataverse is under maintenance, TDC datasets cannot be retrieved. That happens rarely; please check the status on the Dataverse website.
License
The TDC codebase is licensed under the MIT license. For individual dataset usage, please refer to the dataset license on the website.




